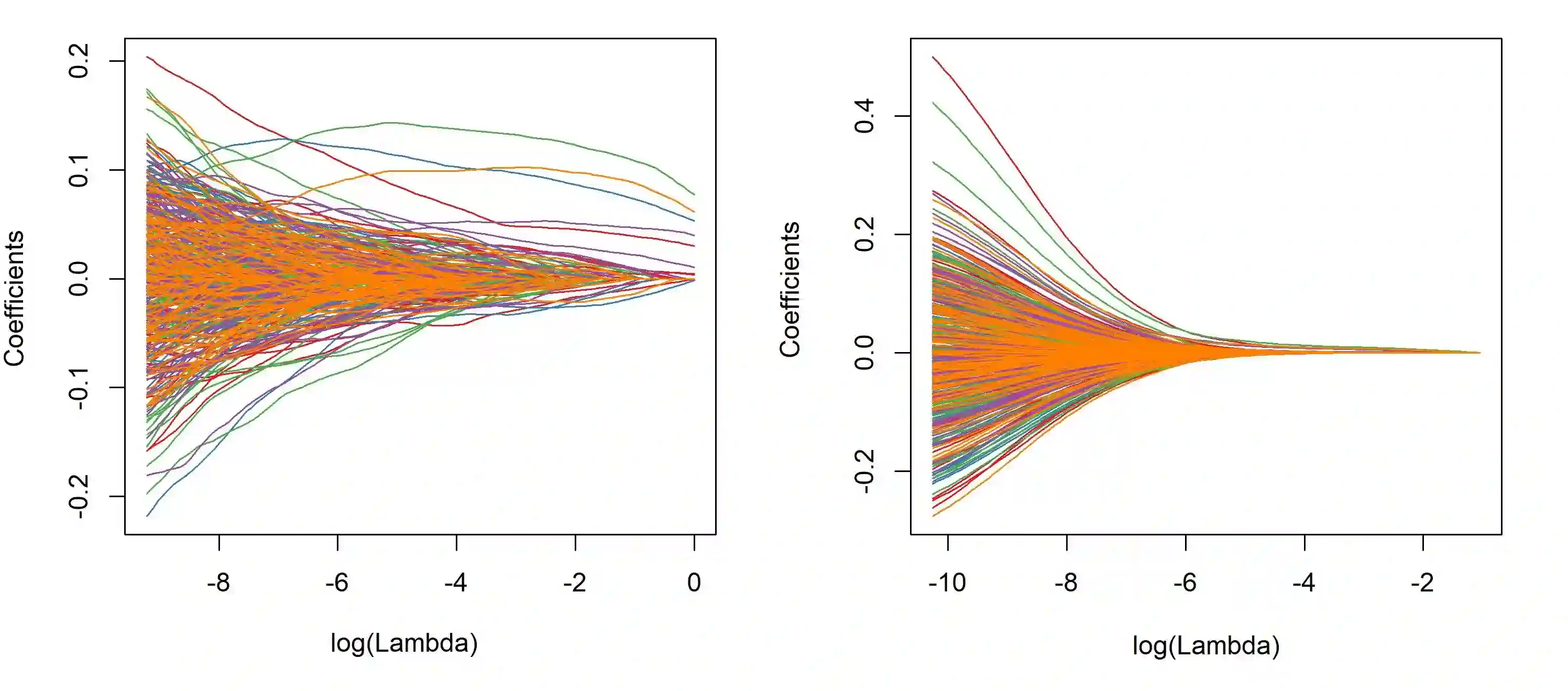
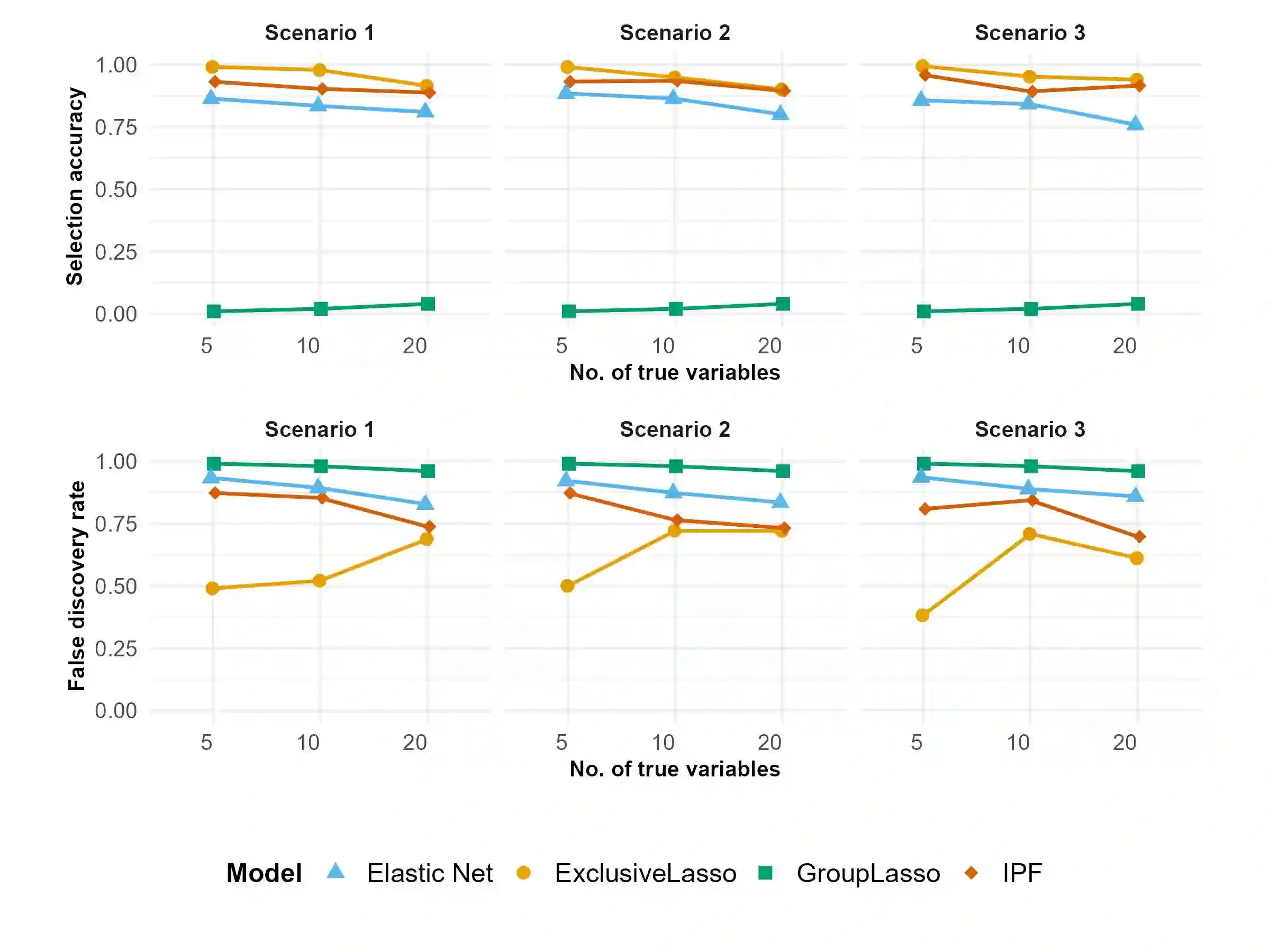
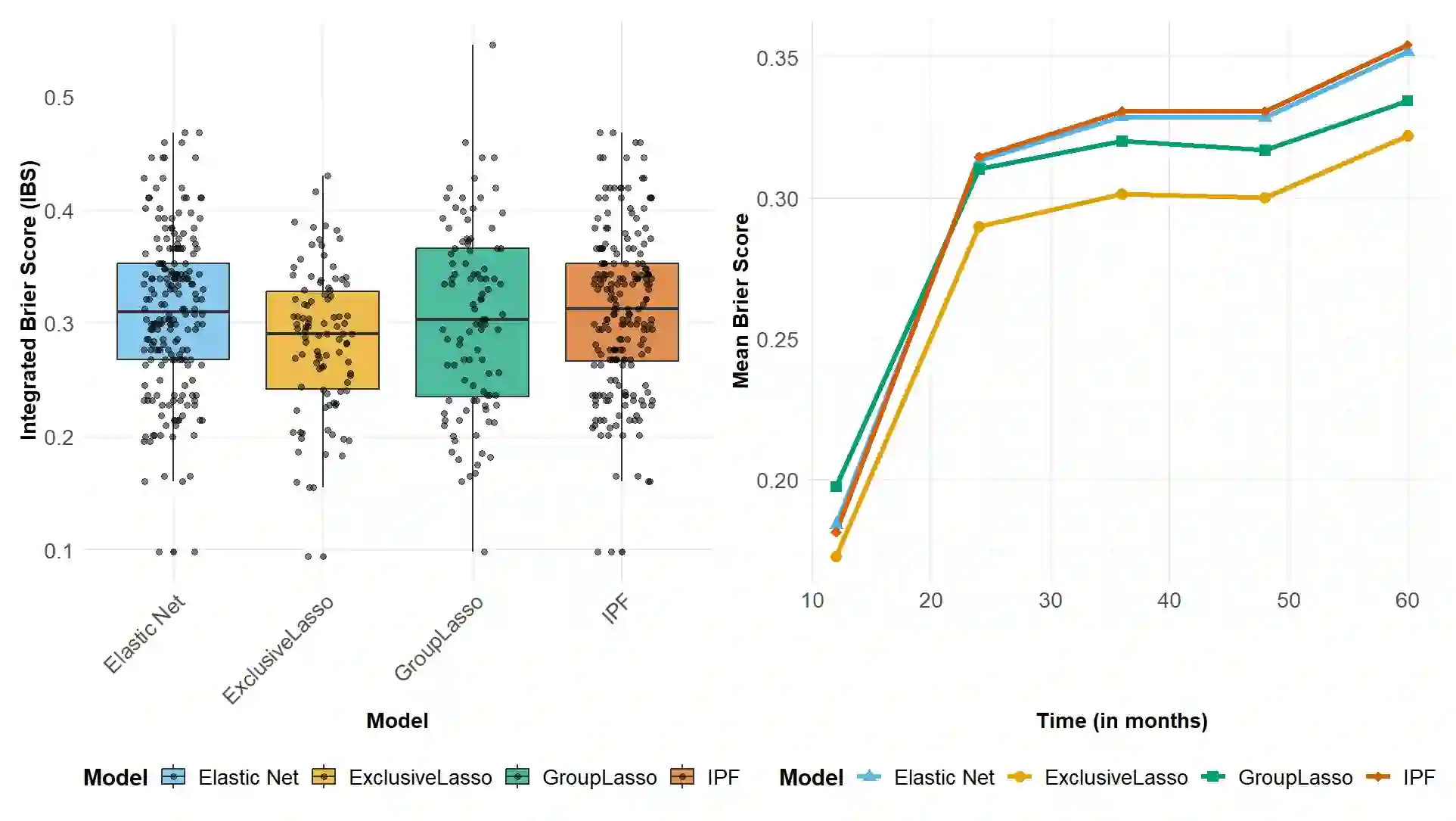
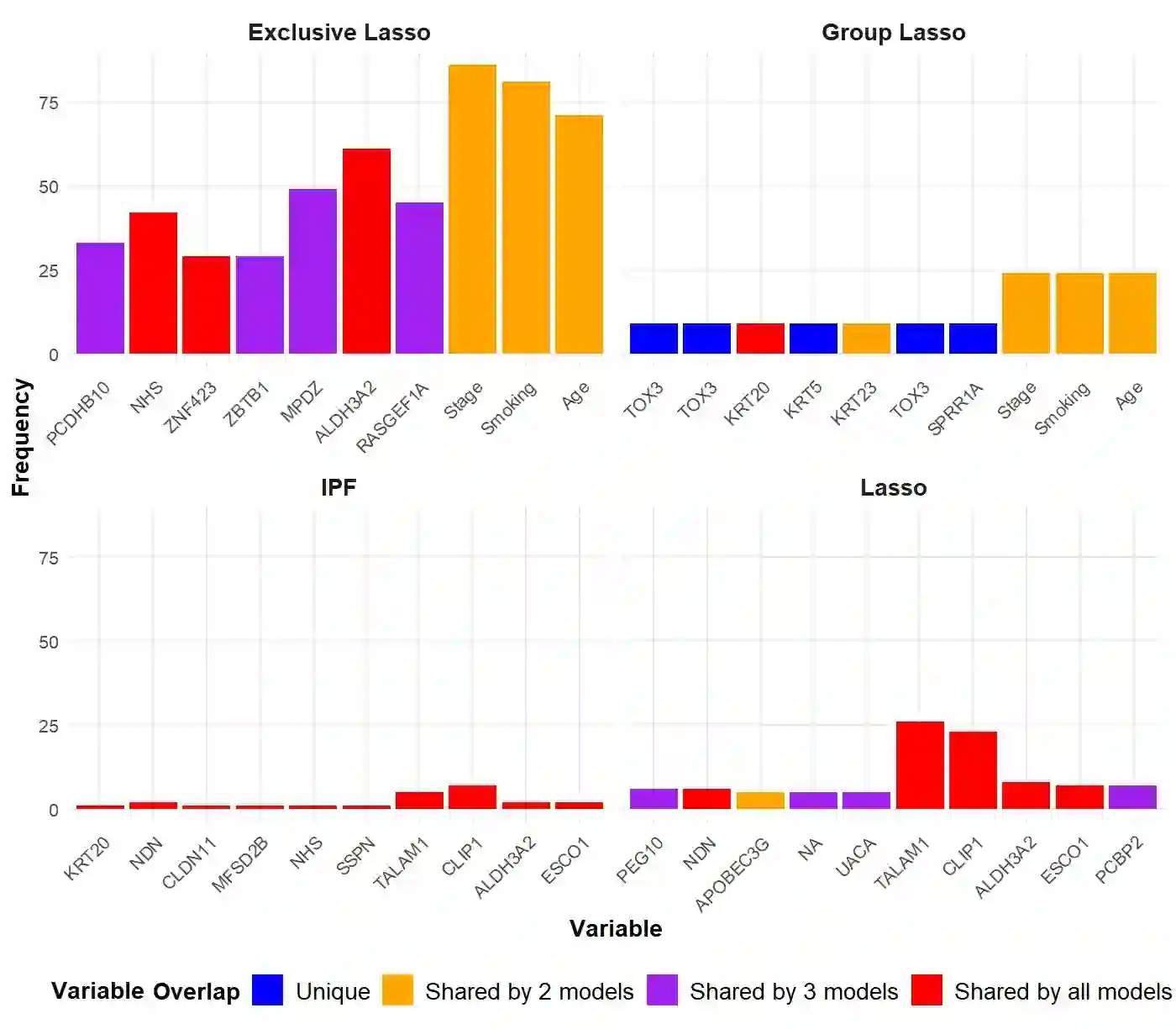
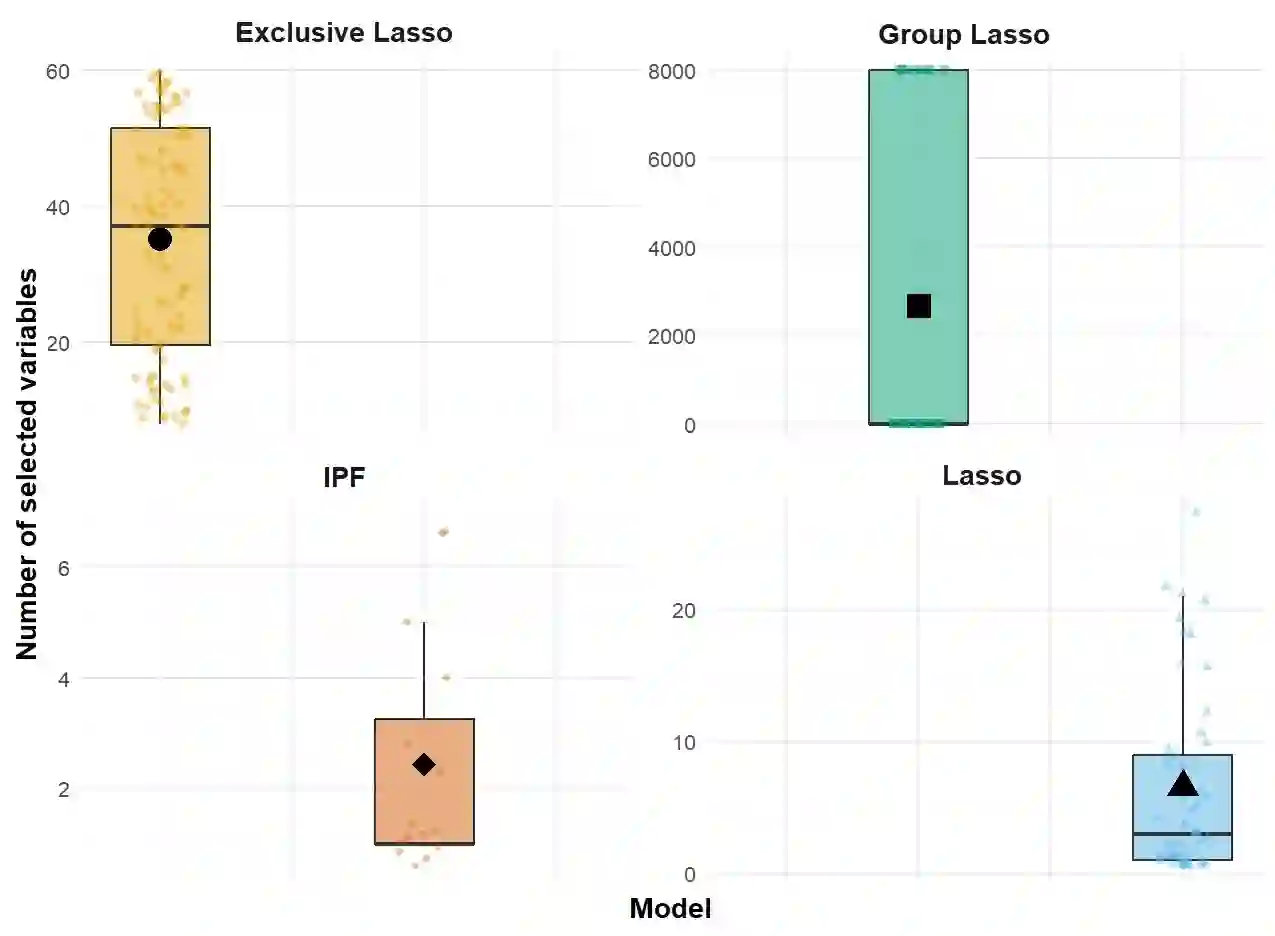
The integration of high-dimensional genomic data and clinical data into time-to-event prediction models has gained significant attention due to the growing availability of these datasets. Traditionally, a Cox regression model is employed, concatenating various covariate types linearly. Given that much of the data may be redundant or irrelevant, feature selection through penalization is often desirable. A notable characteristic of these datasets is their organization into blocks of distinct data types, such as methylation and clinical predictors, which requires selecting a subset of covariates from each group due to high intra-group correlations. For this reason, we propose utilizing Exclusive Lasso regularization in place of standard Lasso penalization. We apply our methodology to a real-life cancer dataset, demonstrating enhanced survival prediction performance compared to the conventional Cox regression model.
翻译:随着高维基因组数据与临床数据集的日益丰富,将这两类数据整合至事件时间预测模型已受到广泛关注。传统方法通常采用Cox回归模型,将各类协变量线性拼接后进行建模。鉴于数据中可能存在大量冗余或无关特征,通过惩罚项进行特征筛选往往是必要的。此类数据集的一个显著特征在于其按不同数据类型(如甲基化数据与临床预测因子)分块组织的结构,由于组内相关性较高,需要从每个组别中选择协变量子集。为此,我们提出采用排他性Lasso正则化替代标准Lasso惩罚方法。我们将所提方法应用于真实癌症数据集,结果表明相较于传统Cox回归模型,该方法能有效提升生存预测性能。