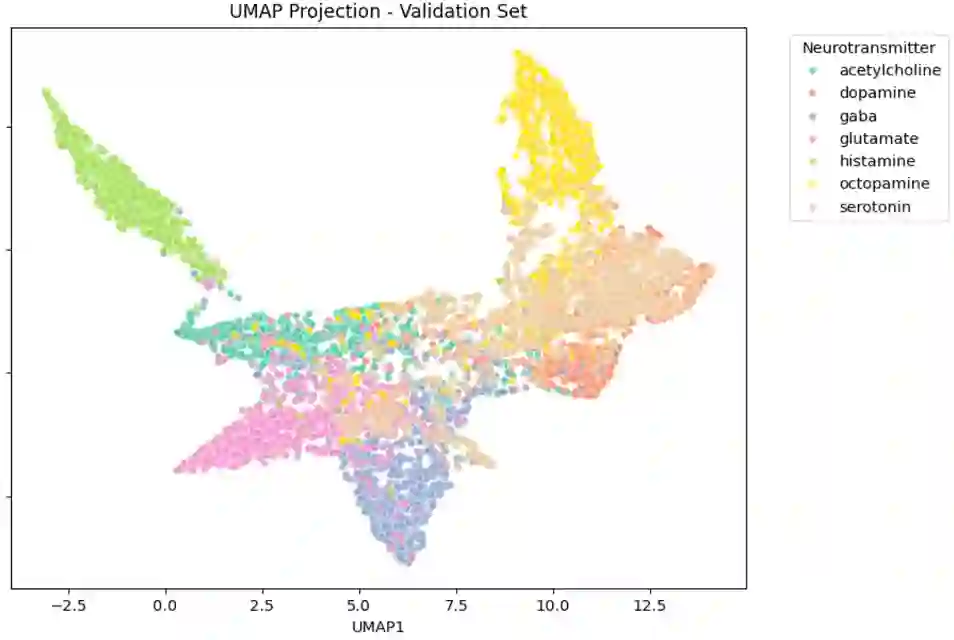
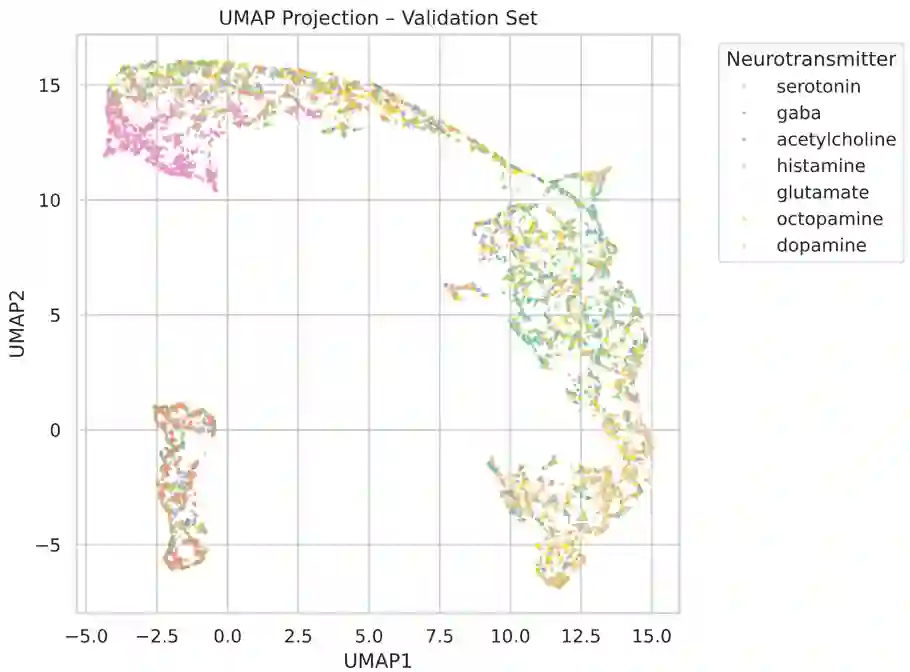
Separating synapses into different classes based on their appearance in EM images has many applications in biology. Examples may include assigning a neurotransmitter to a particular class, or separating synapses whose strength can be modulated from those whose strength is fixed. Traditionally, this has been done in a supervised manner, giving the classification algorithm examples of the different classes. Here we instead separate synapses into classes based only on the observation that nearby synapses in the same neuron are likely more similar than synapses chosen randomly from different cells. We apply our methodology to data from {\it Drosophila}. Our approach has the advantage that the number of synapse types does not need to be known in advance. It may also provide a principled way to select ground-truth that spans the range of synapse structure.
翻译:根据突触在电子显微镜图像中的形态特征将其划分为不同类别,在生物学领域具有广泛的应用价值。例如,可将特定神经递质归属于特定突触类别,或区分强度可调节的突触与强度固定的突触。传统方法通常采用监督学习范式,为分类算法提供不同类别的标注样本。本研究提出一种仅依据"同一神经元内邻近突触的相似度高于随机选取的不同细胞间突触"这一观察的自监督分类方法,并将该方法应用于果蝇实验数据。本方法的优势在于无需预先设定突触类型的数量,同时为构建覆盖突触结构全谱系的基准真值标注提供了理论依据。