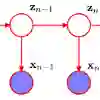
Single nucleotide polymorphism (SNP) datasets are fundamental to genetic studies but pose significant privacy risks when shared. The correlation of SNPs with each other makes strong adversarial attacks such as masked-value reconstruction, kin, and membership inference attacks possible. Existing privacy-preserving approaches either apply differential privacy to statistical summaries of these datasets or offer complex methods that require post-processing and the usage of a publicly available dataset to suppress or selectively share SNPs. In this study, we introduce an innovative framework for generating synthetic SNP sequence datasets using samples derived from time-inhomogeneous hidden Markov models (TIHMMs). To preserve the privacy of the training data, we ensure that each SNP sequence contributes only a bounded influence during training, enabling strong differential privacy guarantees. Crucially, by operating on full SNP sequences and bounding their gradient contributions, our method directly addresses the privacy risks introduced by their inherent correlations. Through experiments conducted on the real-world 1000 Genomes dataset, we demonstrate the efficacy of our method using privacy budgets of $\varepsilon \in [1, 10]$ at $\delta=10^{-4}$. Notably, by allowing the transition models of the HMM to be dependent on the location in the sequence, we significantly enhance performance, enabling the synthetic datasets to closely replicate the statistical properties of non-private datasets. This framework facilitates the private sharing of genomic data while offering researchers exceptional flexibility and utility.
翻译:单核苷酸多态性(SNP)数据集是遗传学研究的基础,但在共享时会带来重大的隐私风险。SNP之间的相关性使得诸如掩码值重构、亲属关系推断和成员推断攻击等强对抗攻击成为可能。现有的隐私保护方法要么对这些数据集的统计摘要应用差分隐私,要么提供复杂的方法,这些方法需要后处理并使用公开可用的数据集来抑制或有选择地共享SNP。在本研究中,我们引入了一种创新的框架,利用时间非齐次隐马尔可夫模型(TIHMM)生成的样本来合成SNP序列数据集。为了保护训练数据的隐私,我们确保每个SNP序列在训练过程中仅贡献有限的影响,从而实现强差分隐私保证。至关重要的是,通过对完整的SNP序列进行操作并限制其梯度贡献,我们的方法直接解决了由其固有相关性引入的隐私风险。通过在真实世界的1000 Genomes数据集上进行的实验,我们证明了我们的方法在$\delta=10^{-4}$、隐私预算为$\varepsilon \in [1, 10]$时的有效性。值得注意的是,通过允许HMM的转移模型依赖于序列中的位置,我们显著提升了性能,使得合成数据集能够紧密复现非隐私数据集的统计特性。该框架促进了基因组数据的隐私共享,同时为研究人员提供了卓越的灵活性和实用性。