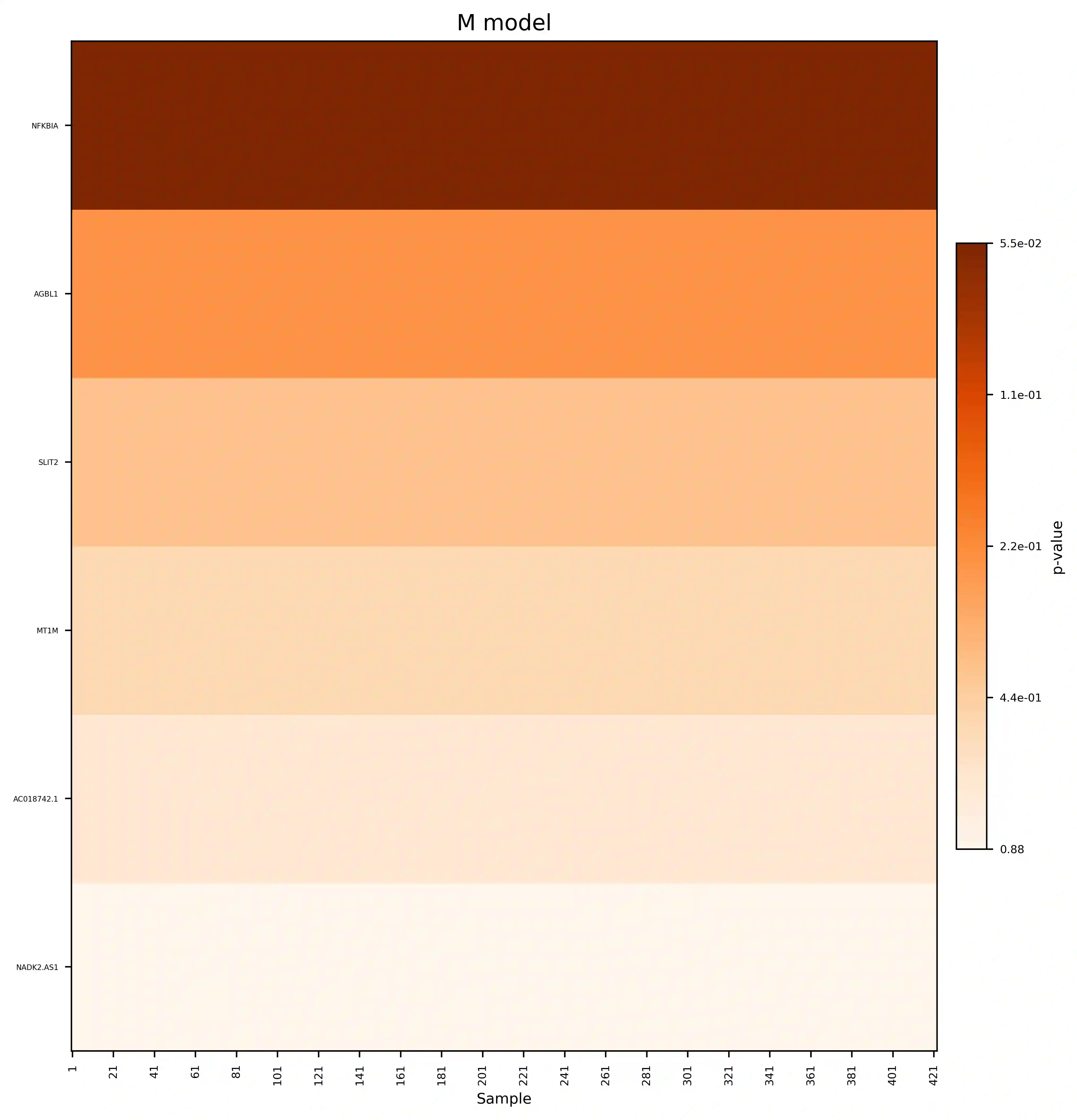
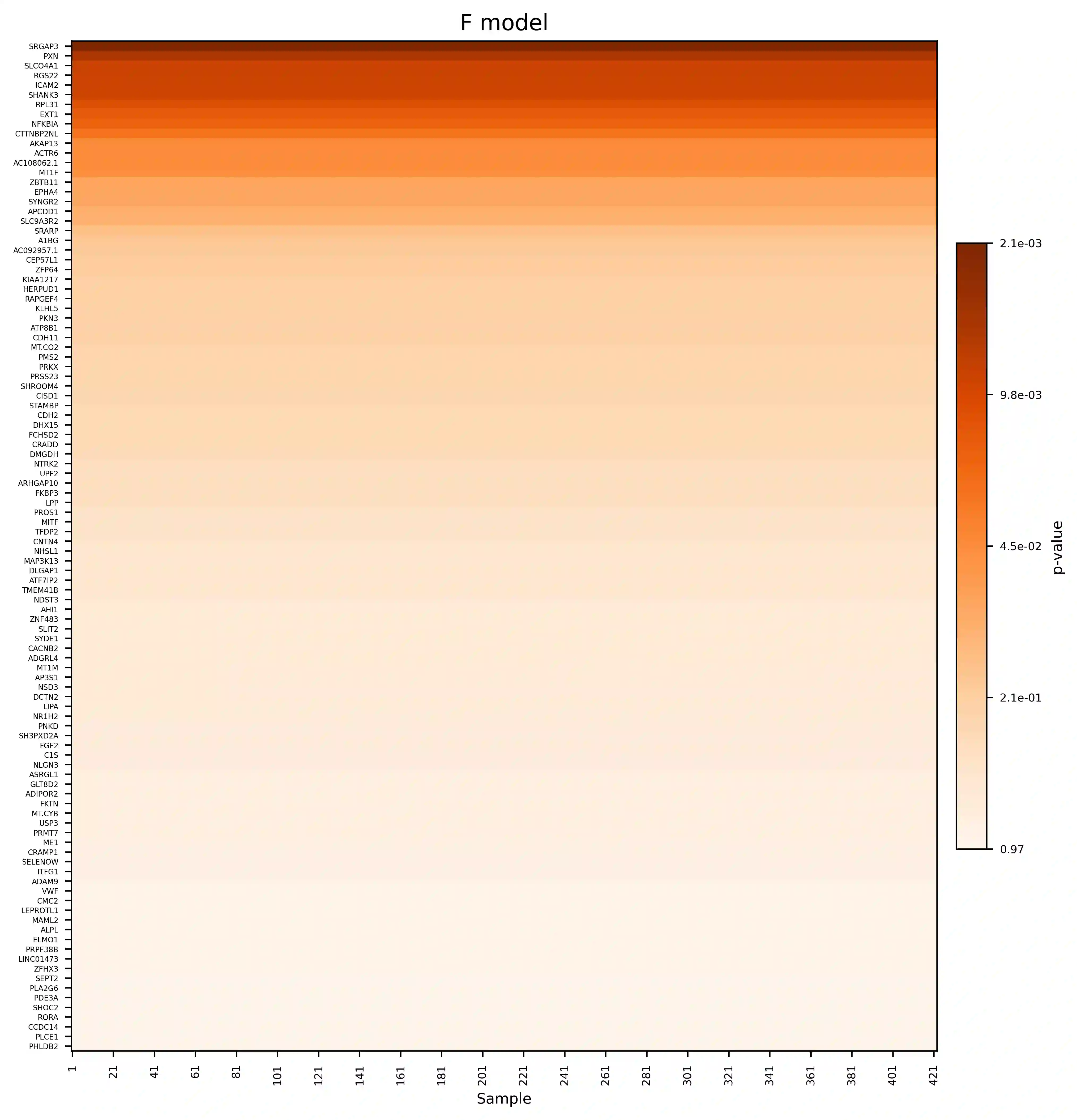
Single-cell RNA sequencing (scRNA-seq) has revolutionized the study of cellular heterogeneity, enabling detailed molecular profiling at the individual cell level. However, integrating high-dimensional single-cell data into causal mediation analysis remains challenging due to zero inflation and complex mediator structures. We propose a novel mediation framework leveraging zero-inflated negative binomial models to characterize cell-level mediator distributions and beta regression for zero-inflation proportions. The model can identify expression level as well as expressed proportion that could mediate disease-leading causal pathway. Extensive simulation studies demonstrate improved power and controlled false discovery rates. We further illustrate the utility of this approach through application to ROSMAP single-cell transcriptomic data, uncovering biologically meaningful mediation effects that enhance understanding of disease mechanisms.
翻译:单细胞RNA测序(scRNA-seq)技术革新了细胞异质性研究,使得在单个细胞水平进行精细分子谱分析成为可能。然而,由于零膨胀现象和复杂的介质结构,将高维单细胞数据整合到因果中介分析中仍面临挑战。我们提出了一种新颖的中介分析框架,该框架利用零膨胀负二项模型刻画细胞水平的中介变量分布,并采用beta回归处理零膨胀比例。该模型能够识别表达水平及表达比例作为疾病发生因果路径中的潜在中介变量。大量模拟研究表明,该方法在提升统计功效的同时能有效控制错误发现率。我们进一步通过应用于ROSMAP单细胞转录组数据展示了该方法的实用性,揭示了具有生物学意义的中介效应,从而深化了对疾病机制的理解。