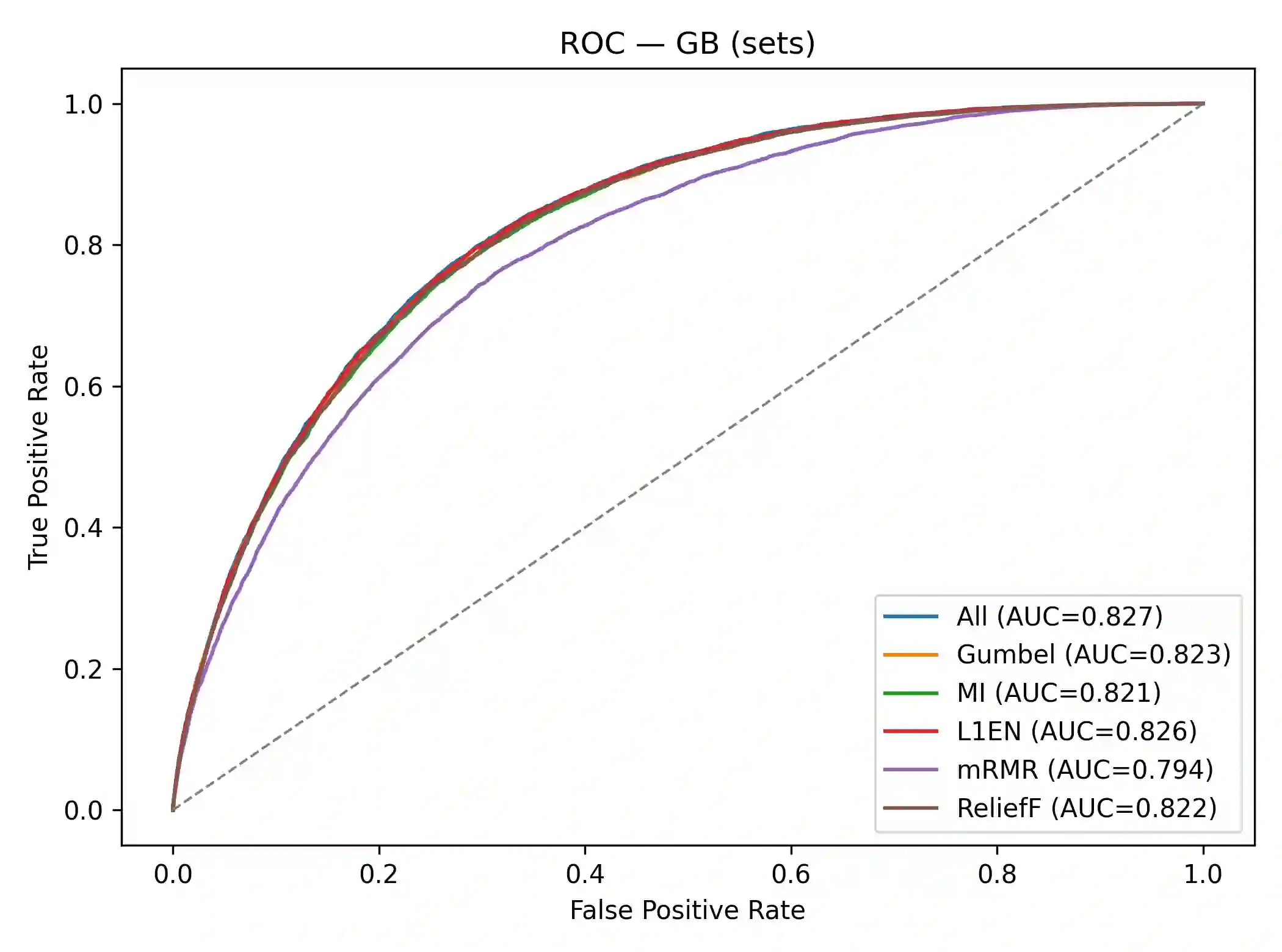
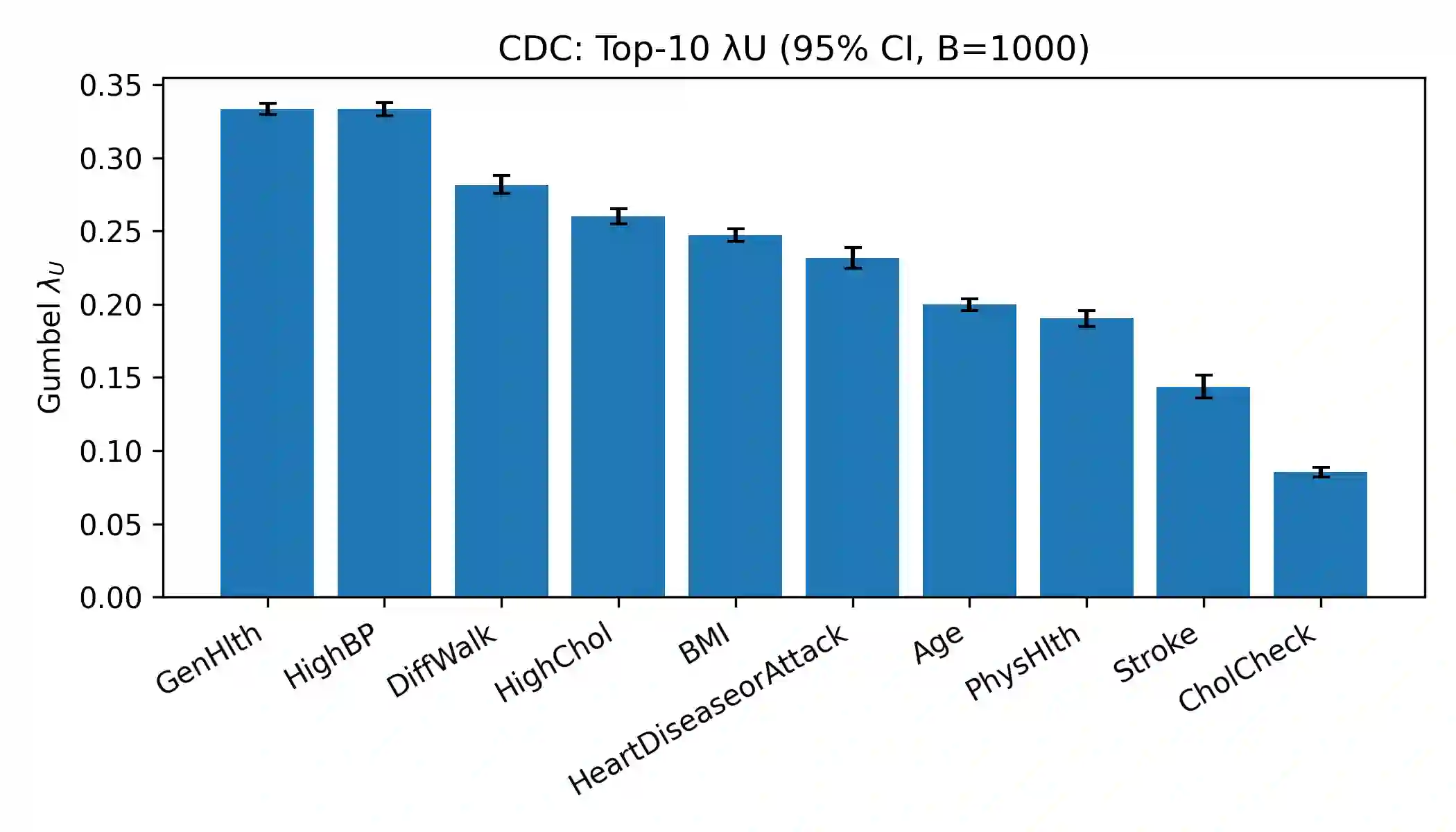
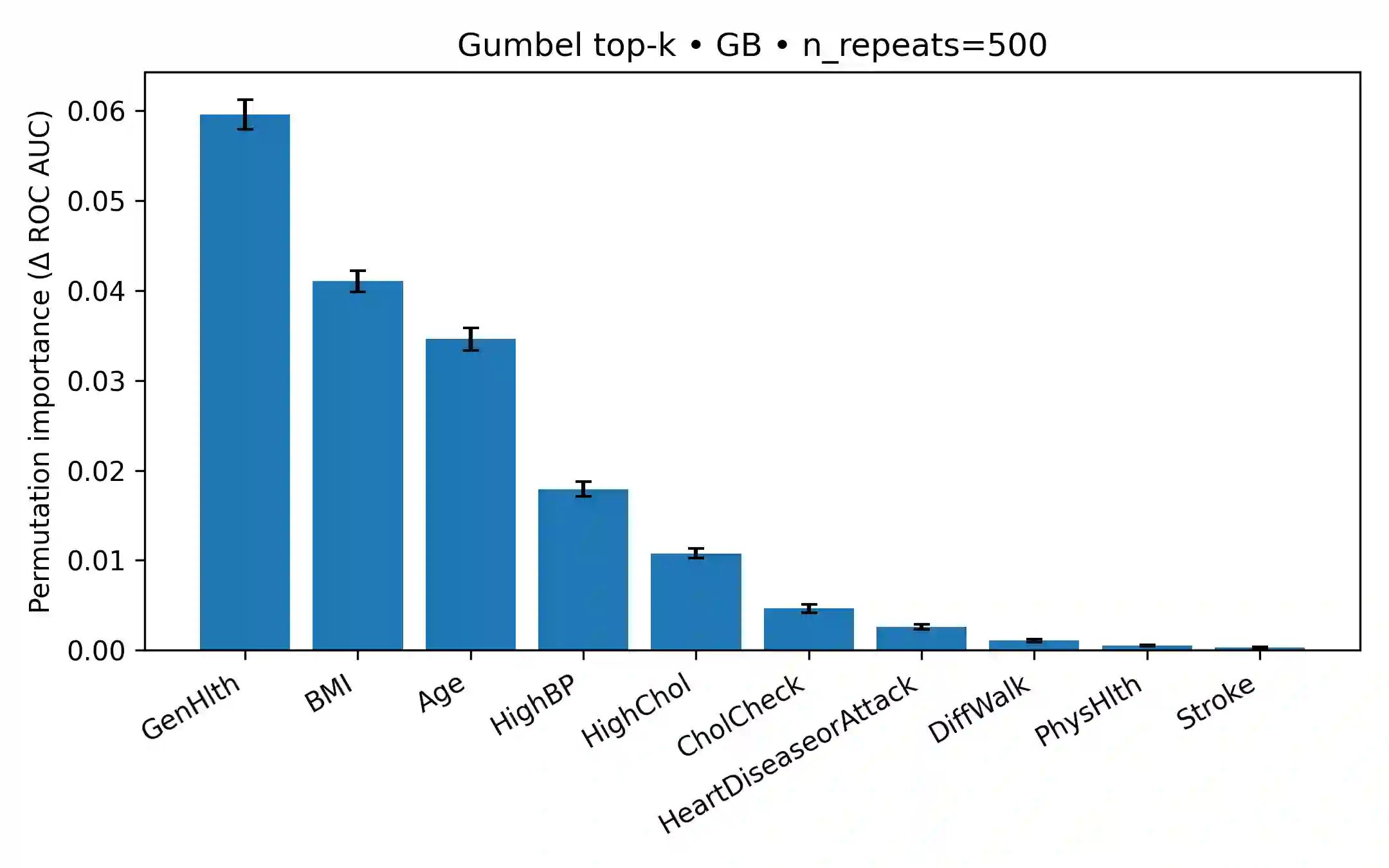
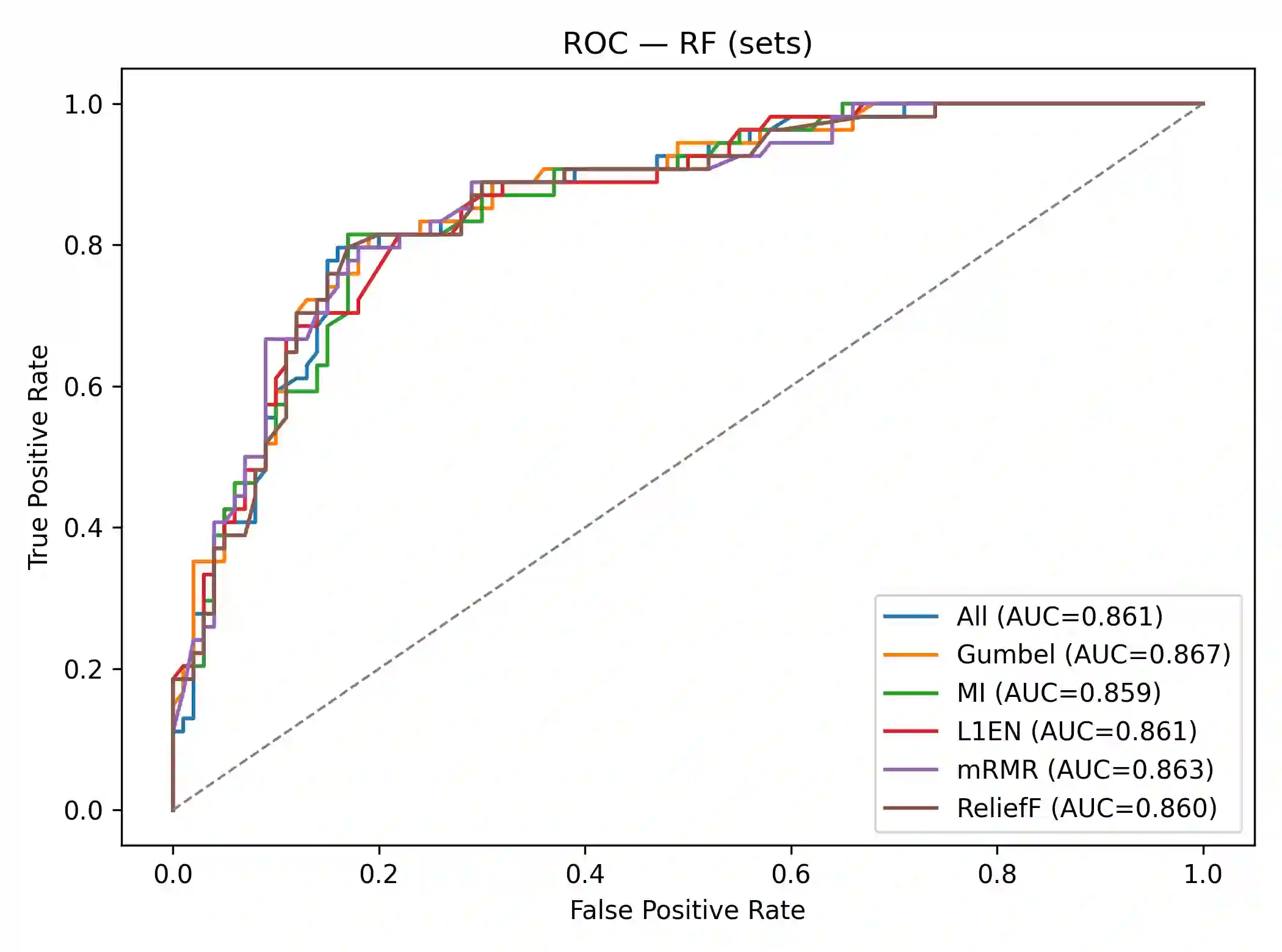
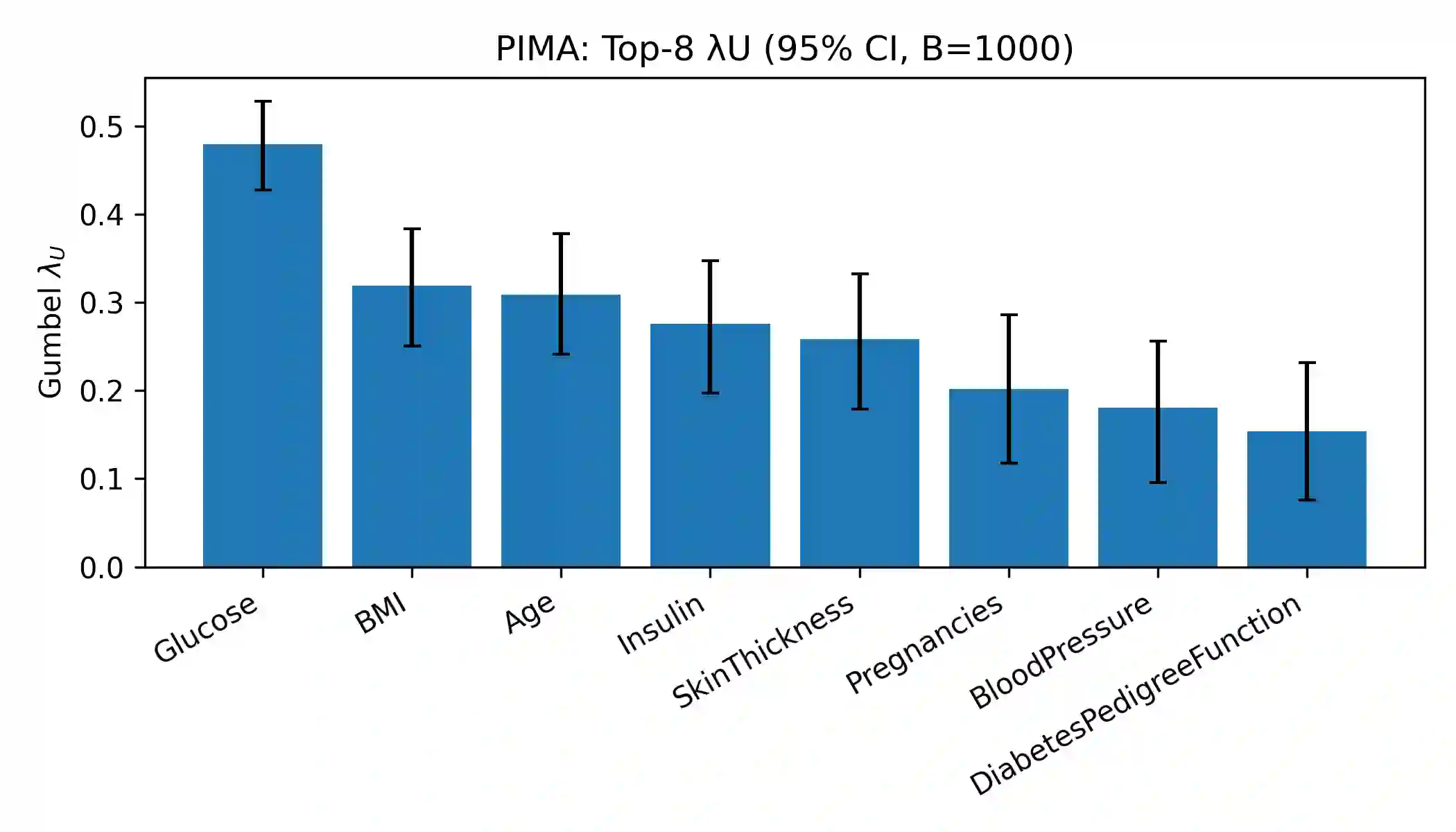
Effective feature selection is vital for robust and interpretable medical prediction, especially for identifying risk factors concentrated in extreme patient strata. Standard methods emphasize average associations and may miss predictors whose importance lies in the tails of the distribution. We propose a computationally efficient supervised filter that ranks features using the Gumbel copula upper tail dependence coefficient ($\lambda_U$), prioritizing variables that are simultaneously extreme with the positive class. We benchmarked against Mutual Information, mRMR, ReliefF, and $L_1$ Elastic Net across four classifiers on two diabetes datasets: a large public health survey (CDC, N=253,680) and a clinical benchmark (PIMA, N=768). Evaluation included paired statistical tests, permutation importance, and robustness checks with label flips, feature noise, and missingness. On CDC, our method was the fastest selector and reduced the feature space by about 52% while retaining strong discrimination. Although using all 21 features yielded the highest AUC, our filter significantly outperformed Mutual Information and mRMR and was statistically indistinguishable from ReliefF. On PIMA, with only eight predictors, our ranking produced the numerically highest ROC AUC, and no significant differences were found versus strong baselines. Across both datasets, the upper tail criterion consistently identified clinically coherent, impactful predictors. We conclude that copula based feature selection via upper tail dependence is a powerful, efficient, and interpretable approach for building risk models in public health and clinical medicine.
翻译:有效的特征选择对于稳健且可解释的医疗预测至关重要,尤其是在识别集中于极端患者分层的风险因素时。标准方法强调平均关联性,可能遗漏那些重要性存在于分布尾部的预测因子。我们提出一种计算高效的监督过滤器,该过滤器使用Gumbel copula上尾相依系数($\lambda_U$)对特征进行排序,优先考虑与阳性类别同时处于极端值的变量。我们在两个糖尿病数据集上,针对四种分类器,与互信息、mRMR、ReliefF以及$L_1$弹性网络进行了基准比较:一个大型公共卫生调查数据集(CDC,N=253,680)和一个临床基准数据集(PIMA,N=768)。评估内容包括配对统计检验、置换重要性,以及针对标签翻转、特征噪声和缺失的稳健性检查。在CDC数据集上,我们的方法是最快的特征选择器,在保留强判别能力的同时,将特征空间减少了约52%。尽管使用全部21个特征获得了最高的AUC,但我们的过滤器显著优于互信息和mRMR,并且与ReliefF在统计上无显著差异。在PIMA数据集上,仅有八个预测因子,我们的排序方法获得了数值上最高的ROC AUC,并且与强基线方法相比未发现显著差异。在两个数据集中,上尾标准一致地识别出临床意义连贯且具有影响力的预测因子。我们得出结论,基于上尾相依性的copula特征选择是构建公共卫生和临床医学风险模型的一种强大、高效且可解释的方法。