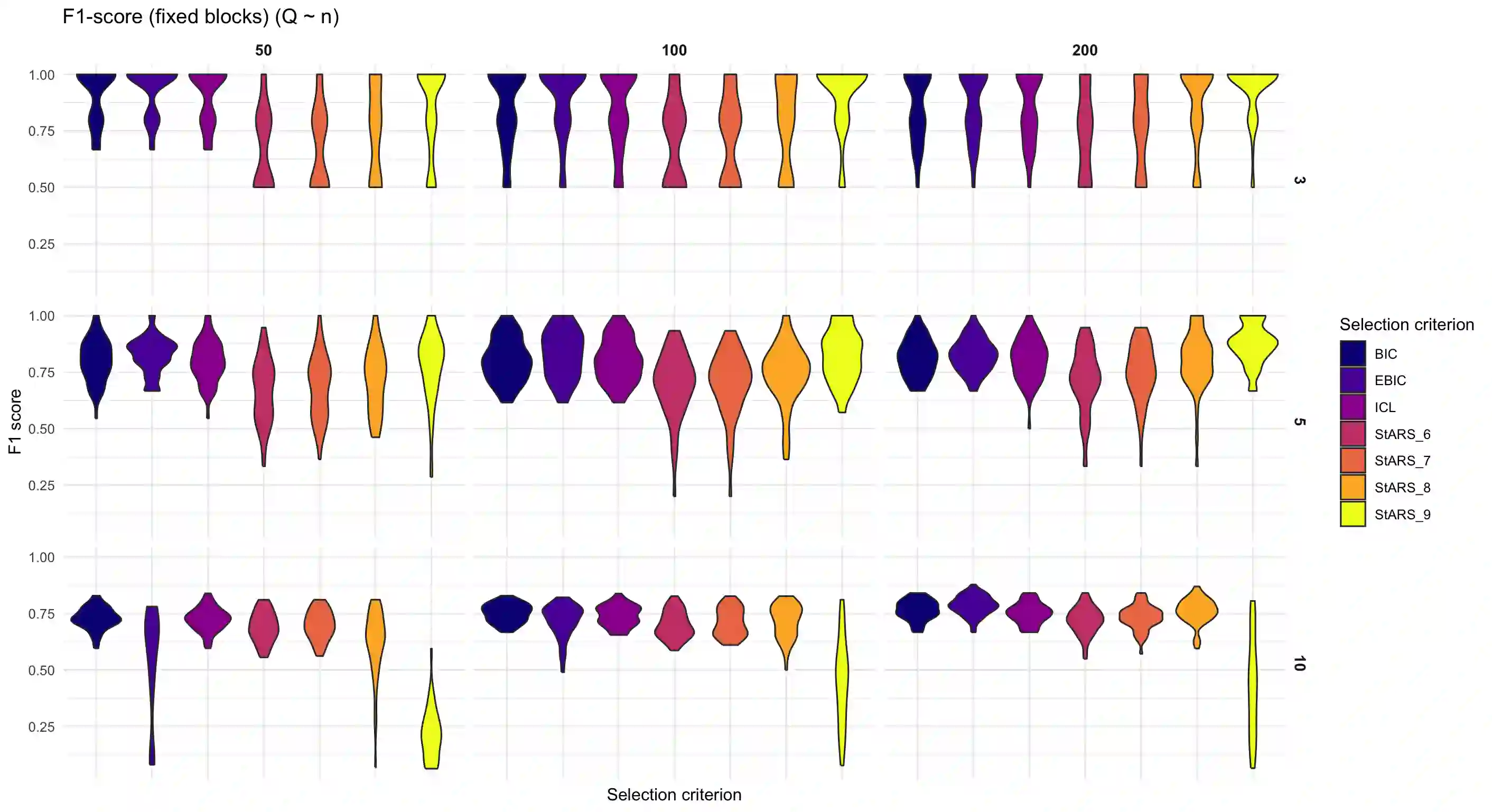
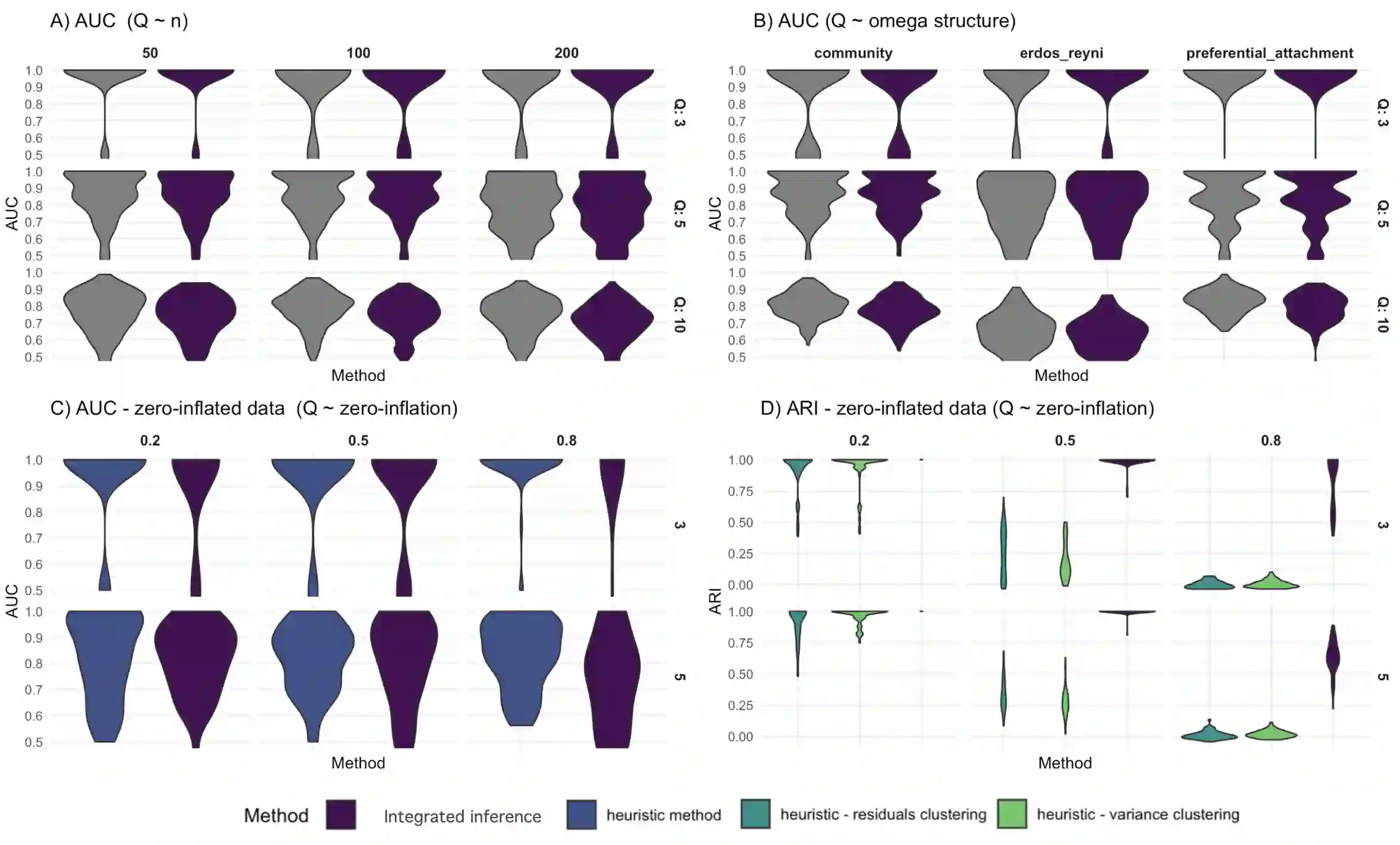
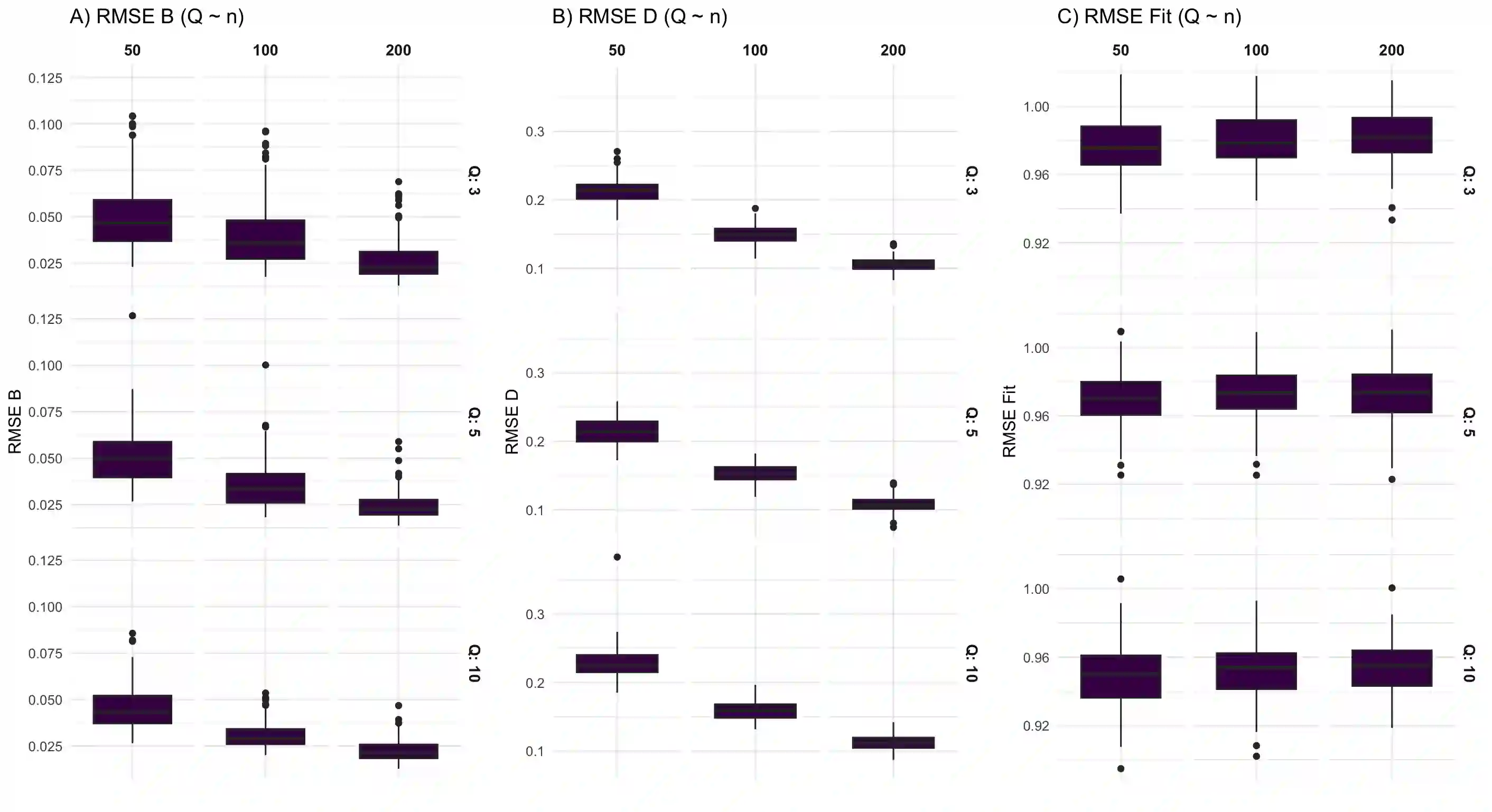
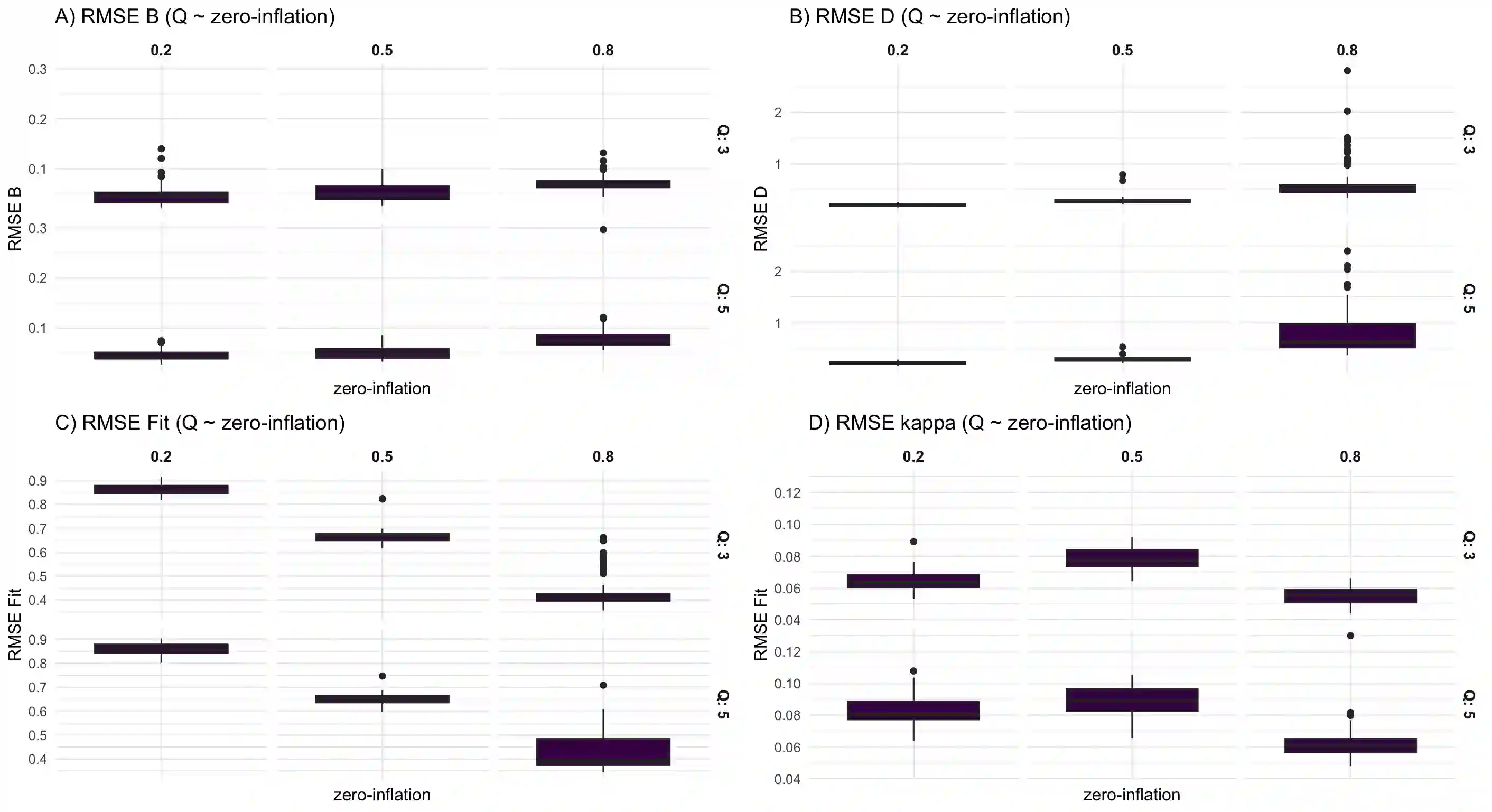
We consider high dimensional Gaussian graphical models inference. These models provide a rigorous framework to describe a network of statistical dependencies between entities, such as genes in genomic regulation studies or species in ecology. Penalized methods, including the standard Graphical-Lasso, are well-known approaches to infer the parameters of these models. As the number of variables in the model (of entities in the network) grow, the network inference and interpretation become more complex. We propose Normal-Block, a new model that clusters variables and consider a network at the cluster level. Normal-Block both adds structure to the network and reduces its size. We build on Graphical-Lasso to add a penalty on the network's edges and limit the detection of spurious dependencies, we also propose a zero-inflated version of the model to account for real-world data properties. For the inference procedure, we propose a direct heuristic method and another more rigorous one that simultaneously infers the clustering of variables and the association network between clusters, using a penalized variational Expectation-Maximization approach. An implementation of the model in R, in a package called normalblockr, is available on github (https://github.com/jeannetous/normalblockr). We present the results in terms of clustering and network inference using both simulated data and various types of real-world data (proteomics, words occurrences on webpages, and microbiota distribution).
翻译:本文研究高维高斯图模型的推断问题。此类模型为描述实体间统计依赖关系的网络提供了严谨的框架,例如基因组调控研究中的基因或生态学中的物种。惩罚方法(包括标准的Graphical-Lasso)是推断这些模型参数的经典方法。随着模型中变量(即网络中的实体)数量的增加,网络推断与解释变得愈发复杂。我们提出Normal-Block这一新模型,该模型对变量进行聚类并在聚类层面构建网络。Normal-Block既能为网络添加结构,又能缩减网络规模。我们在Graphical-Lasso基础上对网络边施加惩罚以限制虚假依赖关系的检测,同时提出模型的零膨胀版本以适应现实数据的特性。针对推断过程,我们提出一种直接启发式方法以及另一种更严谨的方法——后者通过惩罚变分期望最大化方法同步推断变量聚类与聚类间的关联网络。该模型的R语言实现(封装于normalblockr软件包)已在github发布(https://github.com/jeannetous/normalblockr)。我们通过模拟数据和多种现实数据(蛋白质组学、网页词汇共现、微生物群分布)展示了模型在聚类与网络推断方面的结果。