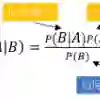
Classifiers assign complex input data points to one of a small number of output categories. For a Bayes classifier whose input space is a graph, we study the structure of the \emph{boundary}, which comprises those points for which at least one neighbor is classified differently. The scientific setting is assignment of DNA reads produced by \NGSs\ to candidate source genomes. The boundary is both large and complicated in structure. We introduce a new measure of uncertainty, Neighbor Similarity, that compares the result for an input point to the distribution of results for its neighbors. This measure not only tracks two inherent uncertainty measures for the Bayes classifier, but also can be implemented for classifiers without inherent measures of uncertainty.
翻译:分类器将复杂的输入数据点划分至少量输出类别之一。针对输入空间为图的贝叶斯分类器,我们研究了其边界结构——该边界由至少存在一个相邻点被分类至不同类别的所有点构成。本研究的科学背景是将二代测序技术产生的DNA读段分配至候选源基因组。该边界不仅规模庞大,且结构复杂。我们提出了一种新的不确定性度量指标——邻域相似度,该指标通过比较输入点的分类结果与其邻域点结果的分布来实现。该度量不仅能追踪贝叶斯分类器两种固有的不确定性指标,还可应用于不具备固有不确定性度量的分类器。